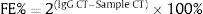IL-6 mRNA expression is significantly high in many autoimmune diseases such as Behçet's disease; this is often related with more aggressive phenotypes. Nevertheless, the essential molecular process for its high expression has not been completely realized. The aim of this study was undertaken to estimate the gene copy number variation and promoter methylation to IL-6's high expression.
MethodsThis study was performed on 51 patients and 61 healthy controls. Initially, DNA and RNA were extracted from all specimens. Promoter methylation levels of IL-6 were evaluated by MeDIP-qPCR technique. Also, IL-6 gene expression was measured by Real-time PCR. After that, we evaluated the relationship between gene expression and methylation, as well as their relationship with clinical specification.
ResultsAs we expected, the expression level of IL-6 gene increased significantly in the patient group compared to the healthy subjects. Also, the relative promoter methylation level of the IL-6 mRNA was significantly lower in patient group compared to healthy group (p<0.001).
DiscussionWe disclosed that the promoter hypomethylation may be considered as one of the main defects for IL-6 mRNA high expression in patients with Behçet's disease.
La expresión de ARNm de IL-6 es significativamente elevada en muchas enfermedades autoinmunes, tales como el síndrome de Behçet, y ello se relaciona a menudo con fenotipos más agresivos. Sin embargo, no se ha comprendido plenamente el proceso molecular esencial para esta expresión elevada. El objetivo de este estudio fue la estimación de la variación del número de copias del gen, y la metilación del promotor de la expresión elevada de IL-6.
MétodosEste estudio se realizó en 51 pacientes y 61 controles sanos. Al inicio, se extrajo ADN y ARN de todas las muestras. Se evaluaron los niveles de metilación del promotor de IL-6 mediante la técnica MeDIP-qPCR. También se midió la expresión del gen IL-6 mediante PCR a tiempo real. Tras ello, evaluamos la relación entre la expresión del gen y la metilación, así como su relación con la especificación clínica.
ResultadosSegún lo previsto, el nivel de expresión del gen IL-6 se incrementó significativamente en el grupo de pacientes, con respecto a los sujetos sanos. También encontramos que el nivel relativo de metilación del promotor de ARNm de IL-6 fue considerablemente menor en el grupo de pacientes, con respecto al grupo sano (p<0,001).
DiscusiónConcluimos que la hipometilación del promotor puede considerarse uno de los defectos principales de la expresión elevada de ARNm de IL-6, en los pacientes con síndrome de Behçet.
Behcet's is an autoinflammatory disease that is defined by Hulusi Behcet in 1937 as an inflammatory process of unknown etiology, characterized by recurrent aphthous stomatitis, uveitis, genital ulcers, and skin lesions.1 Although Behçet's disease (BD) is an extensive and worldwide disease in different parts of the world, it has remarkable local differences, with the maximum of incidences in the Mediterranean, the Middle East, and the Far East which was locally called the Silk Road. Most prevalence of Behçet's disease has been reported in Turkey including 421 people per 105.2 Among all genetic factors, HLA-B51 has been confirmed as the strongest risk factor for BD, which was verified in various populations. More recently studies have disclosed the association of many non-HLA genes with the BD, such as chemokine receptor 1 (CCR1),3 Vitamin D Receptor (VDR),4 interleukin-23 (IL-23),5 IL-23R and IL-12RB2,3,6 fork head box P3 (Foxp3),7 IL-2, IL-4, transforming growth factor (TGF)-beta,8 IL-279 and IL-610 for BD.
IL-6 is a multifunctional cytokine with a wide-range of biologic activities such as the regulation of the acute-phase response to infection.11 Dysregulation and dysfunction of IL-6 gene expression contributes to the inception of numerous diseases, such as several types of cancer, autoimmune disease for example rheumatoid arthritis (RA), type 1 diabetes (T1D), inflammatory bowel disease (IBD), multiple sclerosis (MS), and autoinflammatory disorders such as Behçet's disease (BD).11,12 According to previous studies, it was found that IL-6 exerts its action via the activation of the JAK/STAT (Janus kinase/signal transducer and activator of transcription) and MAPK (mitogenactivated protein kinase) cascades.11,13
Previous studies have found four types of epigenetic mechanisms, which are responsible for regulating the expression of genes including: DNA methylation, histone modification, non-coding RNAs (nc RNAs) and chromatin remodeling.14–16 DNA methylation is an epigenetic alteration that efficiently controls gene expression via gene silencing, and may significantly contribute to the risks of many multiplex diseases, including cancer, autoimmune disease, and metabolic disorders.16–18 Recent researches of cytokine's gene expression have focused on the epigenetic alterations, because the DNA methylation status at CpG sites is often related with cytokine expression changes.19 Decreasing of methylation is related with chromatin's open position, but increasing of methylation corresponds to chromatin's closed status.20 Then, the extent to which CpG islands methylation is associated with the level of cytokine expression. Thus, we questioned whether DNA methylation of IL-6 is reduced in BD T cells with modifying transcription factor recruitment.
Proinflammatory and anti-inflammatory agents are involved in the beginning and development of BD; thus, a comprehensive survey of the regulatory mechanism for the production of cytokines during the progression of BD is the most importance challenge. IL-6 produced mainly by monocytes, B cells, T helper type 2 (Th2) and endothelial cells and plays an essential role in encouraging inflammation and stimulating the immune response.21 Also, the production of IL-6 in the joint tissues is usually done in response to interleukin-1 beta (IL-1β) and tumor necrosis factor alpha (TNF-α) and is mostly performed by chondrocytes, osteoblasts, macrophages, and adipocytes.22,23 Also, IL-6 is an important cytokine in BD pathogenesis as well as it plays a key role in differentiation of CD4+ T cells to Th17 cells.24 IL-6 is an inflammatory cytokine that has been associated in several immune-mediated inflammatory diseases.25 IL-6 can encourage inflammation by increasing the production of inflammatory cytokines such as TNF-α, IL-1β and other agents. In addition, this cytokine plays a key role by inhibiting the anti-inflammatory cytokines and interleukin-1 receptor antagonists (IL-1Ra) production.26,27
Evidence has indicated that IL-6 is involved in the pathogenesis of autoimmune disease. In this study, we analyzed the IL-6 expression in patient with BD in comparison with healthy groups. Also, we evaluated the methylation level of interleukin-6 in both healthy and patient groups.
Methods and materialsPatients and healthy controls study groupAll specimens presented their written informed agreement for this study, and the study protocol was permitted by the ethics committee of Tabriz University of Medical Sciences, Tabriz, Iran (Permit Number: TBZMED.REC.1394.640). The study group consisted of 51 Iranian patients with BD (61.7% men and 38.3% women), range 16–60 years and 61 healthy candidates. The analysis of BD was based on the international study group criteria for BD.28 Features of the patients were evaluated at the time of diagnosis. The control group composed of 61 healthy subjects who were matched with age, gender, and ethnic of patients (59% men versus 41% women). Healthy participants had no clinical or laboratory signs of autoimmune or inflammatory diseases.
DNA, RNA isolation and RT-PCR methodPeripheral blood mononuclear cells (PBMCs) were extracted from EDTA blood tubes by Ficoll (Lymphodex, Inno -Train, Germany) density-gradient centrifugation and directly stored at −80°C until use. Genomic DNA samples of BD and healthy controls were isolated by using the rapid genomic DNA extraction (RGDE) method from the peripheral blood collected in tubes containing EDTA.29 Total RNA was extracted from the PBMCs according to the protocol of TRIzol (Invitrogen, San Diego, CA), followed by reverse transcription using the reverse transcription reagent kit (Thermo Fisher scientific, USA). Then, purity and concentration of total RNA were estimated by nanodrop ND1000 and at 260–280nm purity of RNAs were assessed. The entirety of total RNA was showed by gel electrophoresis of the individual samples on a 1% agarose gel.
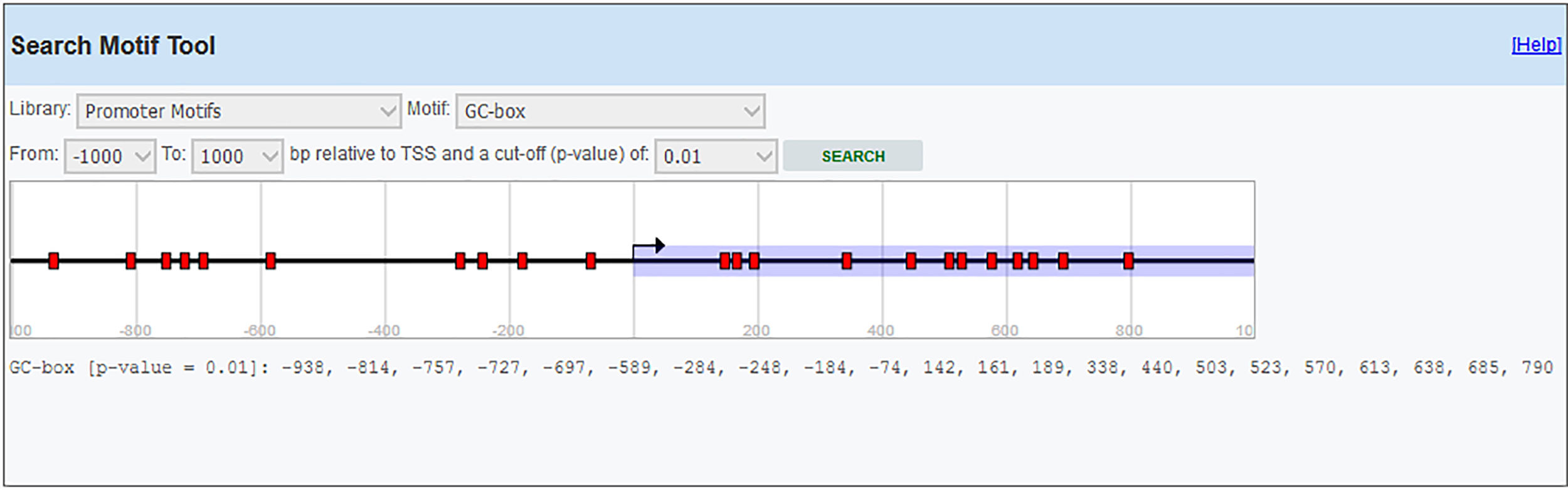
Primer designIL-6 gene sequence and data about promoter were picked up from the National Center for Biotechnology Information (NCBI) and eukaryotic promoter database (EPD). For IL-6 mRNA sequence, the primer pairs were designed using OLIGO7 Software, (Molecular Biology Insights, Inc., Cascade, CO., USA) and MethPrimer online software. (Table 1) Also, IL-6 gene Promoter CpG islands were predicted with eukaryotic promoter database (EPD). One pairs of IL-6 primer were designed using PrimerQuest tool to amplify CpG islands of TSS (transcription start site) upstream (Fig. 1).
Peripheral blood mononuclear cells (PBMCs) were extracted from EDTA blood tubes by Ficoll (Lymphodex, Inno -Train, Germany) density-gradient centrifugation. Total RNA was extracted from the PBMCs using TRIzol (Invitrogen, San Diego, CA), followed by reverse transcription using the reverse transcription reagent kit (thermofisher Scientific, USA) and then the expression of IL-10 was measured by MIC real-time instrument (Bio Molecular Systems, AUSTRALIA). The following sequences of the sense and antisense primers of IL-6 were used: forward 5′-GCAGAAAACAACCTGAACC-3′ and reverse 5′-GCTTGTTCCTCACTACTCTC-3′. β-Actin was chosen as the internal reference for expression and copy number variation detection and its expression was evaluated by the following primers in Table 1. Relative expression levels of IL-6 were calculated using the ΔΔCt formula. All tests were performed in at three biological repeats.
Methylated DNA immunoprecipitation assessmentOne pair of primers were designed using Primer Quest Tool and methMarker (PREMIER Biosoft, CA, USA) to amplify CpG islands of TSS (transcription start site) upstream (Fig. 1). Methylated DNA immunoprecipitation (MeDIP) was performed using EpiQuik™ MeDIP Ultra Kit (Epigentek, Farmingdale, NY, USA). This Kit contains all components essential for doing a successful MeDIP procedure using DNA extracted from PBMCs such as a methylated DNA (mDNA) control and an unmethylated DNA (unDNA) control and control primers that can be used with the control DNA to exhibit the enrichment efficacy and specificity for methylated DNA. The extracted DNA is sonicated to produce random fragments ranging in size from 200 to 800bp using the BANDELIN sonicator (UVV: 3200, Germany) for 15 cycles of 20s on/20s off. Fragment size was confirmed by electrophoresis on a 1.5% agarose gel (Fig. 2). Then, 1μg of DNA is used for following MeDIP enrichment. This method was performed by 5μg of fragmented genomic DNA was diluted to 400μl in TE buffer and DNA was denatured at 95°C for 5min followed by immediate cooling on ice for 5min.
MeDIP-QPCR amplification was carried out using 1μl of eluted DNA in a 20μl PCR reaction along with primer sets. The assays were performed in three replicates and relative methylation fold change was measured for each sample with FE% method which is referred to below. QPCR cycles and the timing steps include an activation cycle, a denaturation cycle at 95°C for 2min and 40 cycles of extension at 95°C for 10s, 54°C for 30s, and finally 72°C for 20s.
The fold enrichment (FE) of IL-6 fragments was calculated using a formula described in manufacture protocol of this kit using a ratio of amplification efficiency of the MeDIP sample over that of non-immune IgG:
Statistical analysisStatistical analysis was performed using SPSS software version 17.0 (SPSS, Chicago, IL, USA). Normal distributions were tested with the Kolmogorov–Smirnov test with Lilliefors correction. Quantitative data were presented as mean±standard deviation (SD). The differences in mRNA and serum levels of IL-6 between control and BD groups were evaluated by Mann–Whitney U test. p-Value <0.05 was considered as significant difference.
ResultsSubject characteristicsPatient's characteristics are shown in Table 2. The patient group consisted of 29 males and 18 females, with a mean age of 38.02±10.25 years. The control subjects included 37 males and 24 females and had a mean age of 37.4±8.5 years. No significant difference was observed in age between patients with BD and controls (p>0.05).
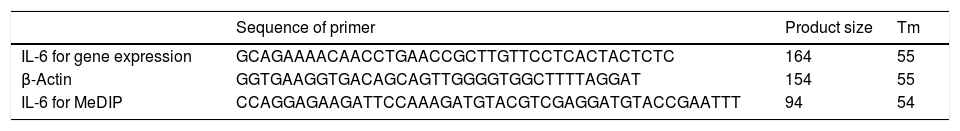
Clinical profile of patients with IL-6 gene expression and its methylation.
| Characteristics and clinical features expression | Frequency | Change fold of IL-6 expression(mean±SD) | p-Value | Methylation level of IL-6 expression(mean±SD) | p-Value |
|---|---|---|---|---|---|
| Age | |||||
| <45 | 31 (63%) | 1.5±1.2 | 0.13 | 0.69±0.09 | 0.76 |
| ≥45 | 16 (28%) | 2.1±1.6 | 0.71±0.06 | ||
| Gender | |||||
| Male | (29) 61.7% | 1.2±1.3 | 0.41 | 0.75±0.1 | 0.18 |
| female | (18) 38.3% | 1.05±1.3 | 0.72±0.08 | ||
| HLA-B5- | |||||
| Positive | 17 (33%) | 1.93±1.32 | 0.36 | 0.68±0.06 | 0.31 |
| Negative | 9 (21%) | 1.35±1.86 | 0.65±0.09 | ||
| HLA-B51 | |||||
| Positive | 8 (15%) | 1.8±1.6 | 0.9 | 0.68±.06 | 0.5 |
| Negative | 7 (13%) | 1.7±1.5 | 0.65±0.07 | ||
| HLA-B27 | |||||
| Positive | 2 (6%) | 0.34±0.46 | 0.17 | 0.69±0.03 | 0.88 |
| Negative | 23 (44%) | 1.88±1.5 | 0.68±0.08 | ||
| Oral aphtha | |||||
| Positive | 45 (96%) | 1.6±1.4 | 0.35 | 0.7±0.08 | 0.5 |
| Negative | 2 (4%) | 2.6±0.87 | 0.74±0.04 | ||
| Genital ulcer | |||||
| Positive | 24 (51%) | 1.7±1.6 | 0.78 | 0.68±0.08 | 0.18 |
| Negative | 23 (49%) | 1.66±1.16 | 0.71±0.07 | ||
| Arthritis | |||||
| Positive | 9 (19%) | 1.7±1.4 | 0.97 | 0.7±0.08 | 0.04 |
| Negative | 38 (81%) | 1.7±1.17 | 0.74±0.05 | ||
| Sever B.D. | |||||
| Positive | 30 (64%) | 1.5±1.5 | 0.41 | 0.69±0.08 | 0.41 |
| Negative | 17 (36%) | 1.91±1.1 | 0.71±0.08 | ||
| Severe eye involvement | |||||
| Positive | 10 (22%) | 1.7±1 | 0.9 | 0.7±0.06 | 0.64 |
| Negative | 36 (78%) | 1.7±1.5 | 0.7±0.08 | ||
| EN | |||||
| Positive | 7 (16%) | 1.5±1.5 | 0.62 | 0.7±0.12 | 0.94 |
| Negative | 38 (84%) | 1.8±1.2 | 0.7±0.07 | ||
| Phlebitis | |||||
| Positive | 6 (11%) | 2.7±0.97 | 0.04 | 0.74±0.05 | 0.29 |
| Negative | 41 (89%) | 1.6±1.3 | 0.7±0.08 | ||
| Ocular | |||||
| No eye involvement | 11 (23%) | 1.7±0.8 | 0.44 | 0.75±0.1 | 0.26 |
| One eye activity | 15 (33%)) | 2.1±1.2 | 0.67±0.07 | ||
| Bilateral activity | 21 (44%) | 1.4±1.7 | 0.7±0.07 | ||
| Cataract | |||||
| Positive | 9 (19%) | 2.1±1.1 | 0.28 | 0.69±0.07 | 0.84 |
| Negative | 36 (74%) | 1.6±1.4 | 0.7±0.08 | ||
| Vision loss | |||||
| One eye | 6 (13%) | 1.69±1.2 | 0.84 | 0.67±0.08 | 0.45 |
| No eye | 38 (77%) | 1.78±1.34 | 0.7±0.08 | ||
As shown in the table, items that have a statistically significant difference are shown as bold. IL-6: interleukin-6, SD: standard deviation, HLA: Human leukocyte antigen, BD: Behçet's disease, EN: Erythema Nodosum.
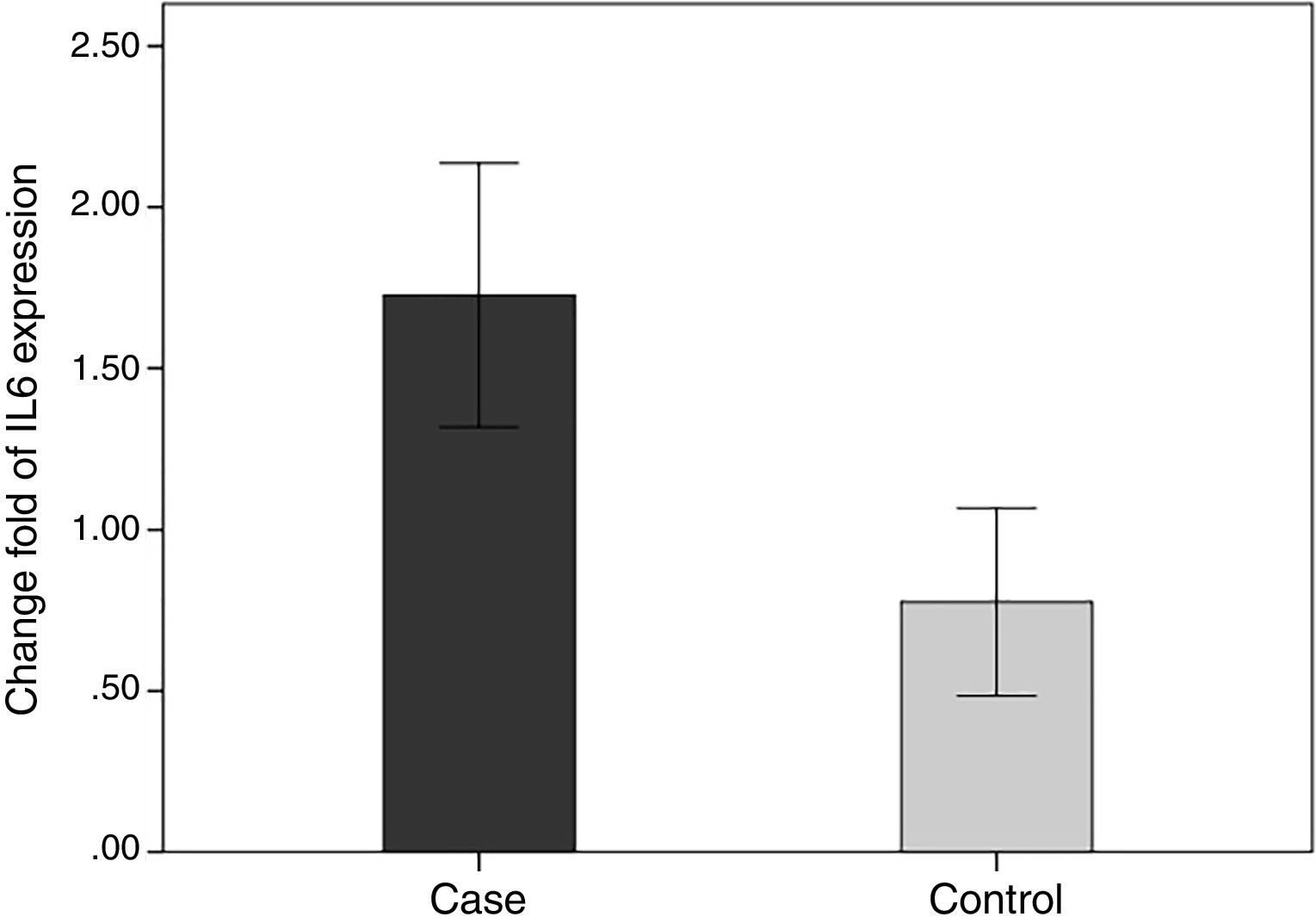
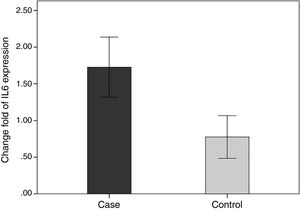
In order to compare the level of interleukin-6 (IL-6) gene expression in two groups of BD and healthy subjects, an independent T-test was adopted, taking into consideration the data obtained by employing Kolmogorov–Smirnov test, was normal (p-value >0.05). The obtained results were indicative of significant difference between two groups (p-value <0.05). As we expected, the level of gene expression showed increase in BD individuals group in comparison with healthy group (Fig. 3). Also in the patient groups, we analyzed the relationship between gene expression and clinical features. As the results indicated, the gene expression level of IL-6 was significantly different in phlebitis section that is shown as bold in table. Interleukin-6 gene expression was also significantly increased in individuals with positive phlebitis (p-value <0.05) (Table 2).
Change fold of IL-6 expression. Regarding the average changes in the expression of the IL-6 gene in the patients and the healthy groups, the amount of it is comparable to that of the healthy group in the patient group, which indicates that IL-6 gene expression was increased among the patients in the patient group.
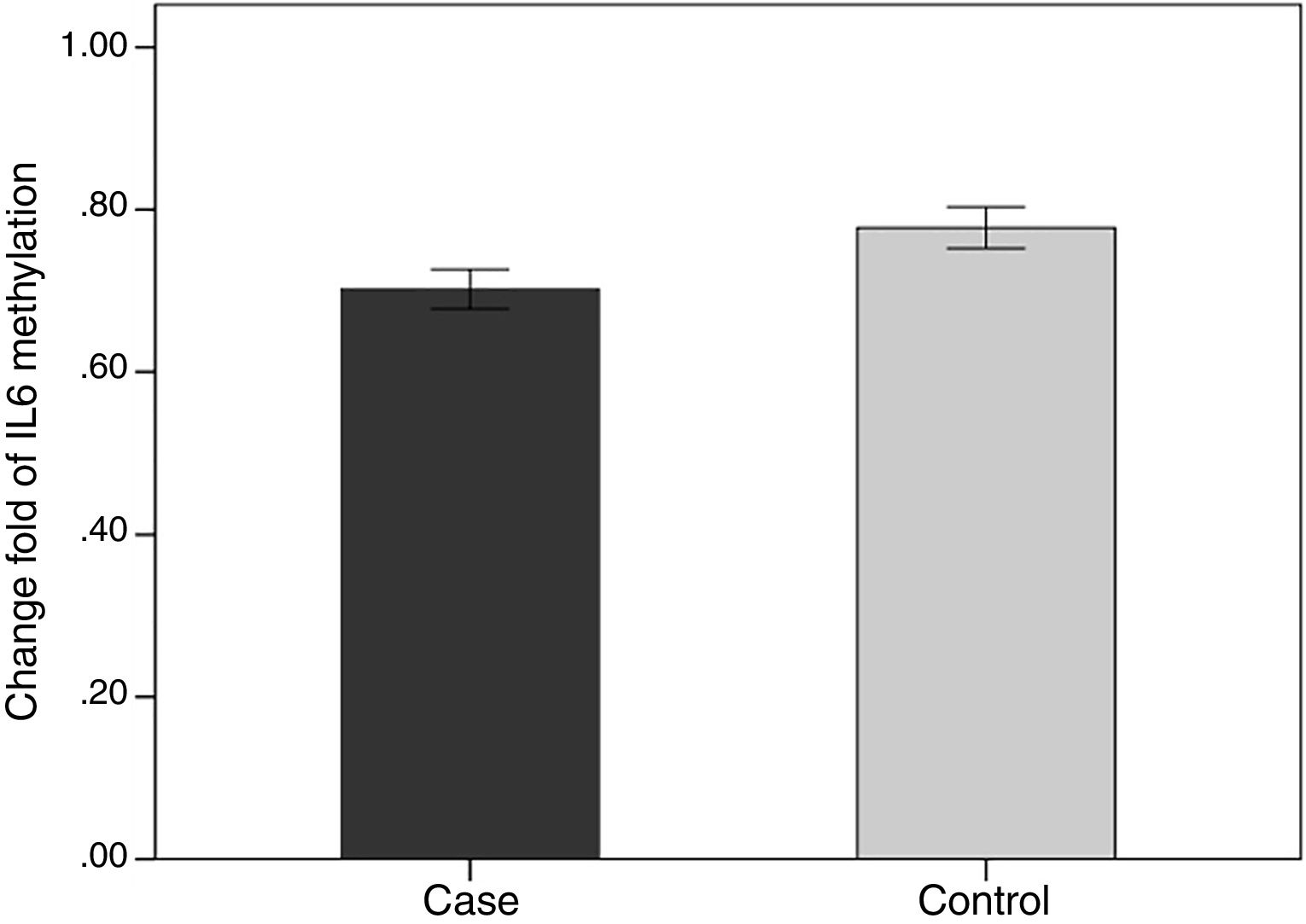
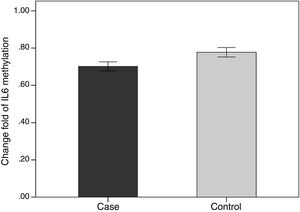
We evaluated the association between the methylation pattern of the IL-6 gene and mRNA expression. The mRNA (1.7±1.3 versus 0.77±1.14, p<0.05) of IL6 was significantly higher in the patients with BD than healthy groups. Also, in order to compare the methylation status of interleukin-6 (IL-6 in two groups of BD and healthy individuals, an independent T-test was adopted, taking into consideration the data obtained by employing Kolmogorov–Smirnov test, was normal (p-value >0.05). The results showed that there was a statistical difference between both groups in terms of methylation rate of interleukin-6 gene (p-value <0.001). In line with our expectations, the methylation ratio of IL-6 revealed decrease in BD individuals group in comparison with healthy group (0.7±0.08 versus 0.77±0.09) (Fig. 4).
Also in the patient groups, we analyzed the relationship between methylation level and clinical features. As the results showed, the methylation levels had significant differences in the arthritis section. In addition, the methylation level of IL-6 was lower in positive group of arthritis than the negative value (p-value <0.05) (Table 2).
DiscussionBehcet's disease (BD) is an autoinflammatory disease of unknown etiology. Cytokines involved can be classified as Th1, Th2, Th17, Treg types, chemokines and others proinflammatory agents. The cytokine network plays an important role in the pathogenesis of diseases, especially autoimmune disorders, as well in the formation and progression of lesions of the organism and may be blocked by one of the basic links of cytokines, which in turn may obtain the critical clues to view the target therapy program of cytokine in BD.30
Chronic inflammation and responses creates a main mechanism for autoimmune disease. The definite pathways by which the inflammation causes autoimmune are not completely recognized. IL-6 is a multifunctional cytokine produced and released by inflammatory cells and can trigger intracellular transcription factors leading to gene expression, cell division, apoptosis processes and inflammation response.25,31,32 Persistent differences in the production of IL-6 have been identified among different individuals in various populations with polymorphism at the end of 5′ region33 and the genetic associations of IL-6 polymorphism have been identified with rates of infections, inflammation and immune diseases. This suggested that the production rate of this cytokine is a risk factor for most diseases and their condition.34 However, to date, the role of epigenetic variable has not been reported in IL-6 gene on Behçet's disease.
There are some findings indicated that IL-6 may stimulate immune response through the effect on the expression of effective genes by altering patterns of DNA methylation.35 On the other hand, IL-6 production is regulated by methylation of the IL-6 gene promoter.35 DNA demethylation is a property of transcriptionally active genes, and our results are fit with the view that the level of methylation is essential in the regulation of the IL-6 production in patients with BD and could participate in production of this cytokine as seen in BD and other autoimmune disease.35,36
In a study of 51 BD patients aged 16–60y, we examined the level of promoter methylation of IL-6 in association with demographic and clinical risk factors such as gender and severe BD as well as inflammation markers. As the results showed, the gene expression level of IL-6 was significantly different in phlebitis section. IL-6 gene expression was also significantly increased in individuals with positive phlebitis. Furthermore, in order to compare the methylation status of IL-6 in two groups of BD and healthy individuals, the results showed that there was a statistical significant difference between both groups in terms of methylation rate of IL-6 gene (p-value <0.001). According to our results, there was no relationship between the gene expression and its methylation in different parts of the clinical profile. One of the possible reasons for these results is that our population sample size was small, and if we selected a larger population size, our results may be more meaningful.
Epigenetic differences, especially the level of IL-6 methylation between BD patients and healthy group, can be an initial manifestation suggested an increased risk of BD prevalence with the low methylation level of IL-6 promoter. On the other hand, this difference can be considered as a main manipulation for BD prevention or treatment.
Scientists from China reported in previous studies that a significantly higher level of IL-6 in cerebrospinal fluid (CSF) had been detected in Chinese patients with active BD with central nervous system (CNS) involvement.37 Also, in a previous study about the methylation level of the IL-6 promoter in patients with RA in comparison with healthy groups, the results indicated that a single CpG site at -1,181 was considerably less methylated in subjects with rheumatoid arthritis (RA).35 Some pioneering studies demonstrated that dysregulation of immune system is related to aging. Furthermore, these results revealed that the level of IL-6 expression was intensified with age in animals model such as mice.38 It has also been shown that the level of IL-6 are much higher in serum, synovial fluid and synovial tissue in patients with severe RA compared with subjects without inflammatory arthritis which indicates that it is related to the activity of disease.39 Some contradictory results reported in previous studies. A decrease or no effect of age on IL-6 production had been shown already.40 According to the results of this clinical study, significant difference could not be seen between patients when dichotomized by age. These diverse results may be caused by the differences in experimental conditions and the health status of subjects in various investigations.
These results recommend that the level of methylation in promoter region of gene may be a key mechanism for regulation of IL-6 gene expression in BD and IL-6 CpG island hypomethylation might be involved in the progression of BD. Besides, according to the results, molecular methods such as the analysis of DNA methylation could recognize in early detection by changes in the genotype and genome sequencing. This may be used for early diagnosis and initiation of rapid therapies. Therefore, IL-6 can be used as a molecular marker for the diagnosis of Behcet's patients.
ConclusionRegarding the results of expression and methylation of IL-6 in this study, it may be considered that methylation may be as one of the regulatory mechanisms involved in regulating gene expression. Generally, epigenetic alteration such as hypermethylation of IL-6 gene may be a potent therapeutic approach in controlling BD.
Conflict of interestThe authors declare that they have no conflicts of interest.